-Search query
-Search result
Showing 1 - 50 of 125 items for (author: kim & kw)
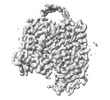
EMDB-35377: 
Cryo-EM structure of GPR156 of GPR156-miniGo-scFv16 complex (local refine)
Method: single particle / : Shin J, Park J, Cho Y

EMDB-35378: 
Cryo-EM structure of miniGo-scFv16 of GPR156-miniGo-scFv16 complex (local refine)
Method: single particle / : Shin J, Park J, Cho Y

EMDB-35380: 
Cryo-EM structure of GPR156-miniGo-scFv16 complex
Method: single particle / : Shin J, Park J, Cho Y

EMDB-35382: 
Cryo-EM structure of GPR156A/B of G-protein free GPR156 (local refine)
Method: single particle / : Shin J, Park J, Cho Y

EMDB-35389: 
Cryo-EM structure of GPR156C/D of G-protein free GPR156 (local refine)
Method: single particle / : Shin J, Park J, Cho Y

EMDB-35390: 
Cryo-EM structure of G-protein free GPR156
Method: single particle / : Shin J, Park J, Cho Y

PDB-8ieb: 
Cryo-EM structure of GPR156 of GPR156-miniGo-scFv16 complex (local refine)
Method: single particle / : Shin J, Park J, Cho Y

PDB-8iec: 
Cryo-EM structure of miniGo-scFv16 of GPR156-miniGo-scFv16 complex (local refine)
Method: single particle / : Shin J, Park J, Cho Y

PDB-8ied: 
Cryo-EM structure of GPR156-miniGo-scFv16 complex
Method: single particle / : Shin J, Park J, Cho Y

PDB-8iei: 
Cryo-EM structure of GPR156A/B of G-protein free GPR156 (local refine)
Method: single particle / : Shin J, Park J, Cho Y

PDB-8iep: 
Cryo-EM structure of GPR156C/D of G-protein free GPR156 (local refine)
Method: single particle / : Shin J, Park J, Cho Y

PDB-8ieq: 
Cryo-EM structure of G-protein free GPR156
Method: single particle / : Shin J, Park J, Cho Y

EMDB-34155: 
Designed pH-responsive P22 VLP
Method: single particle / : Kim KJ, Kim G, Bae JH, Song JJ, Kim HS

PDB-8gn5: 
Designed pH-responsive P22 VLP
Method: single particle / : Kim KJ, Kim G, Bae JH, Song JJ, Kim HS

EMDB-18941: 
SARS-CoV-2 S (Spike) protein (BA.1) in complex with VHH Ma16B06 (sub-volume of two adjacent RBD-VHH modules)
Method: single particle / : Guttler T, Aksu M, Gorlich D

EMDB-28281: 
Subtomogram average of the T4SS of Coxiella Burnetii at pH 4.75
Method: subtomogram averaging / : Kaplan M, Ghosal D
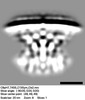
EMDB-28282: 
Subtomogram average of T4SS of Coxiella burnetii at pH 7
Method: subtomogram averaging / : Kaplan M, Shepherd DC, Vankadari N, Kim KW, Larson CL, Przemyslaw D, Beare PA, Krzymowski E, Heinzen RA, Jensen GJ, Ghosal D
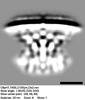
EMDB-28283: 
Subtomogram average of T4SS of Coxiella burnetii at pH7 with an inner membrane mask
Method: subtomogram averaging / : Kaplan M, Shepherd DC, Vankadari N, Kim KW, Larson CL, Przemyslaw D, Beare PA, Krzymowski E, Heinzen RA, Jensen GJ, Ghosal D

EMDB-34813: 
Structure of transforming growth factor beta induced protein (TGFBIp) G623R fibril
Method: helical / : Low JYK, Pervushin K

PDB-8hia: 
Structure of transforming growth factor beta induced protein (TGFBIp) G623R fibril
Method: helical / : Low JYK, Pervushin K

EMDB-33021: 
Cryo-EM structure of the human TRPC5 ion channel in lipid nanodiscs, class1
Method: single particle / : Won J, Jeong H, Lee HH

EMDB-34300: 
Cryo-EM structure of the human TRPC5 ion channel in lipid nanodiscs, class2
Method: single particle / : Won J, Jeong H, Lee HH

EMDB-34301: 
Cryo-EM structure of the human TRPC5 ion channel in complex with G alpha i3 subunits, class2
Method: single particle / : Won J, Jeong H, Lee HH

PDB-7x6c: 
Cryo-EM structure of the human TRPC5 ion channel in lipid nanodiscs, class1
Method: single particle / : Won J, Jeong H, Lee HH

PDB-8gvw: 
Cryo-EM structure of the human TRPC5 ion channel in lipid nanodiscs, class2
Method: single particle / : Won J, Jeong H, Lee HH

PDB-8gvx: 
Cryo-EM structure of the human TRPC5 ion channel in complex with G alpha i3 subunits, class2
Method: single particle / : Won J, Jeong H, Lee HH

EMDB-33022: 
Cryo-EM structure of the human TRPC5 ion channel in complex with G alpha i3 subunits, class1
Method: single particle / : Won J, Jeong H, Lee HH

PDB-7x6i: 
Cryo-EM structure of the human TRPC5 ion channel in complex with G alpha i3 subunits, class1
Method: single particle / : Won J, Jeong H, Lee HH

EMDB-26735: 
Hantavirus ANDV Gn(H) protein in complex with 2 Fabs ANDV-5 and ANDV-34
Method: single particle / : Binshtein E, Crowe JE
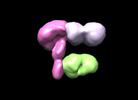
EMDB-26736: 
Hantavirus MAPV Gn(H)/Gc protein in complex with 2 Fabs SNV-24 and SNV-53
Method: single particle / : Binshtein E, Crowe JE

EMDB-27318: 
CryoEM structure of Hantavirus ANDV Gn(H) protein complex with 2Fabs ANDV-5 and ANDV-34
Method: single particle / : Binshtein E, Crowe JE

PDB-8dbz: 
CryoEM structure of Hantavirus ANDV Gn(H) protein complex with 2Fabs ANDV-5 and ANDV-34
Method: single particle / : Binshtein E, Crowe JE
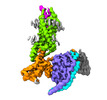
EMDB-33984: 
Complex structure of Neuropeptide Y Y2 receptor in complex with PYY(3-36) and Gi
Method: single particle / : Kang H, Park C, Kim J, Choi HJ

EMDB-33985: 
Complex structure of Neuropeptide Y Y2 receptor in complex with NPY and Gi
Method: single particle / : Kang H, Park C, Kim J, Choi HJ

EMDB-35494: 
Complex structure of Neuropeptide Y Y2 receptor in complex with NPY and Gi (Consensus map)
Method: single particle / : Kang H, Park C, Kim J, Choi HJ

EMDB-35495: 
Complex structure of Neuropeptide Y Y2 receptor in complex with NPY and Gi (Focused map on NPY-Y2R)
Method: single particle / : Kang H, Park C, Kim J, Choi HJ

EMDB-35496: 
Complex structure of Neuropeptide Y Y2 receptor in complex with NPY and Gi (Focused map on Gi-scFv16)
Method: single particle / : Kang H, Park C, Kim J, Choi HJ

EMDB-35497: 
Complex structure of Neuropeptide Y Y2 receptor in complex with PYY(3-36) and Gi (Consensus map)
Method: single particle / : Kang H, Park C, Kim J, Choi HJ

EMDB-35498: 
Complex structure of Neuropeptide Y Y2 receptor in complex with PYY(3-36) and Gi (Focused map on PYY(3-36)-Y2R)
Method: single particle / : Kang H, Park C, Kim J, Choi HJ

EMDB-35499: 
Complex structure of Neuropeptide Y Y2 receptor in complex with PYY(3-36) and Gi (Focused map on Gi-scFv16)
Method: single particle / : Kang H, Park C, Kim J, Choi HJ

PDB-7yon: 
Complex structure of Neuropeptide Y Y2 receptor in complex with PYY(3-36) and Gi
Method: single particle / : Kang H, Park C, Kim J, Choi HJ

PDB-7yoo: 
Complex structure of Neuropeptide Y Y2 receptor in complex with NPY and Gi
Method: single particle / : Kang H, Park C, Kim J, Choi HJ

EMDB-27232: 
Subtomogram of Fluad vaccine hemagglutinin spiked nanodisc
Method: electron tomography / : Gallagher JR, Audray AK

EMDB-27233: 
Tomogram of Flublok vaccine hemagglutinin starfish complexes
Method: electron tomography / : Gallagher JR, Audray AK
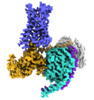
EMDB-24378: 
5-HT2AR bound to a novel agonist in complex with a mini-Gq protein and an active-state stabilizing single-chain variable fragment (scFv16) obtained by cryo-electron microscopy (cryoEM)
Method: single particle / : Barros-Alvarez X, Kim K, Panova O, Roth BL, Skiniotis G

PDB-7ran: 
5-HT2AR bound to a novel agonist in complex with a mini-Gq protein and an active-state stabilizing single-chain variable fragment (scFv16) obtained by cryo-electron microscopy (cryoEM)
Method: single particle / : Barros-Alvarez X, Kim K, Panova O, Roth BL, Skiniotis G

EMDB-31559: 
Cryo-EM structure of BsClpP-ADEP1 complex at pH 6.5
Method: single particle / : Kim L, Lee BG, Kim MK, Kwon DH, Kim H, Brotz-Oesterhelt H, Roh SH, Song HK

EMDB-31560: 
Cryo-EM structure of apo BsClpP at pH 6.5
Method: single particle / : Kim L, Lee BG, Kim MK, Kwon DH, Kim H, Brotz-Oesterhelt H, Roh SH, Song HK

EMDB-31561: 
Cryo-EM structure of BsClpP-ADEP1 complex at pH 4.2
Method: single particle / : Kim L, Lee BG, Kim MK, Kwon DH, Kim H, Brotz-Oesterhelt H, Roh SH, Song HK

EMDB-31562: 
Cryo-EM structure of apo BsClpP at pH 4.2
Method: single particle / : Kim L, Lee BG, Kim MK, Kwon DH, Kim H, Brotz-Oesterhelt H, Roh SH, Song HK
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
